Note
Click here to download the full example code
Within Session SSVEP#
This Example show how to perform a within-session SSVEP analysis on the MAMEM dataset 3, using a CCA pipeline.
The within-session evaluation assesses the performance of a classification pipeline using a 5-fold cross-validation. The reported metric (here, accuracy) is the average of all fold.
# Authors: Sylvain Chevallier <sylvain.chevallier@uvsq.fr>
#
# License: BSD (3-clause)
import warnings
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.pipeline import make_pipeline
import moabb
from moabb.datasets import MAMEM3
from moabb.evaluations import WithinSessionEvaluation
from moabb.paradigms import SSVEP
from moabb.pipelines import SSVEP_CCA
warnings.simplefilter(action="ignore", category=FutureWarning)
warnings.simplefilter(action="ignore", category=RuntimeWarning)
moabb.set_log_level("info")
Loading Dataset#
Load 2 subjects of MAMEM3 dataset
subj = [1, 3]
dataset = MAMEM3()
dataset.subject_list = subj
Choose Paradigm#
We select the paradigm SSVEP, applying a bandpass filter (3-15 Hz) on the data and we keep only the first 3 classes, that is stimulation frequency of 6.66, 7.50 and 8.57 Hz.
Create Pipelines#
Use a Canonical Correlation Analysis classifier
interval = dataset.interval
freqs = paradigm.used_events(dataset)
pipeline = {}
pipeline["CCA"] = make_pipeline(SSVEP_CCA(interval=interval, freqs=freqs, n_harmonics=3))
Get Data (optional)#
To get access to the EEG signals downloaded from the dataset, you could use dataset.get_data(subjects=[subject_id]) to obtain the EEG under MNE format, stored in a dictionary of sessions and runs. Otherwise, paradigm.get_data(dataset=dataset, subjects=[subject_id]) allows to obtain the EEG data in scikit format, the labels and the meta information. In paradigm.get_data, the EEG are preprocessed according to the paradigm requirement.
# sessions = dataset.get_data(subjects=[3])
# X, labels, meta = paradigm.get_data(dataset=dataset, subjects=[3])
Evaluation#
The evaluation will return a DataFrame containing a single AUC score for each subject and pipeline.
overwrite = True # set to True if we want to overwrite cached results
evaluation = WithinSessionEvaluation(
paradigm=paradigm, datasets=dataset, suffix="examples", overwrite=overwrite
)
results = evaluation.process(pipeline)
print(results.head())
MAMEM3-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
No hdf5_path provided, models will not be saved.
MAMEM3-WithinSession: 50%|##### | 1/2 [00:14<00:14, 14.76s/it]/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 5 events (all good), 1 – 4 s (baseline off), ~236 kB, data loaded,
'6.66': 3
'7.50': 0
'8.57': 2>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:279: UserWarning: warnEpochs <Epochs | 4 events (all good), 1 – 4 s (baseline off), ~194 kB, data loaded,
'6.66': 1
'7.50': 0
'8.57': 3>
warn(f"warnEpochs {epochs}")
No hdf5_path provided, models will not be saved.
MAMEM3-WithinSession: 100%|##########| 2/2 [00:29<00:00, 15.03s/it]
MAMEM3-WithinSession: 100%|##########| 2/2 [00:29<00:00, 14.99s/it]
score time samples subject ... channels n_sessions dataset pipeline
0 0.688889 0.046543 45.0 1 ... 14 1 MAMEM3 CCA
1 0.266667 0.046096 45.0 3 ... 14 1 MAMEM3 CCA
[2 rows x 9 columns]
Plot Results#
Here we plot the results, indicating the score for each subject
plt.figure()
sns.barplot(data=results, y="score", x="session", hue="subject", palette="viridis")
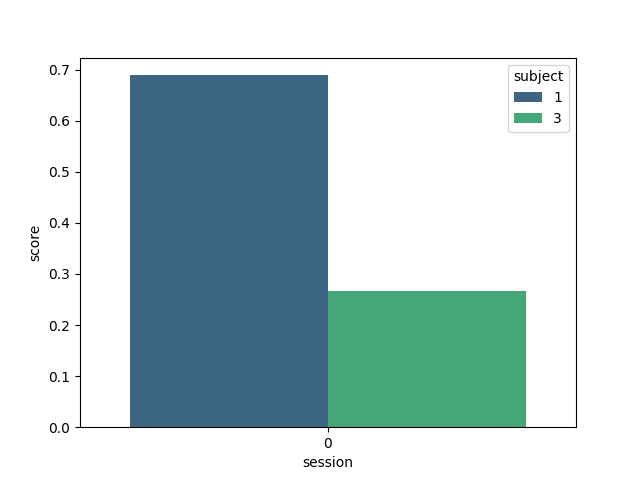
<Axes: xlabel='session', ylabel='score'>
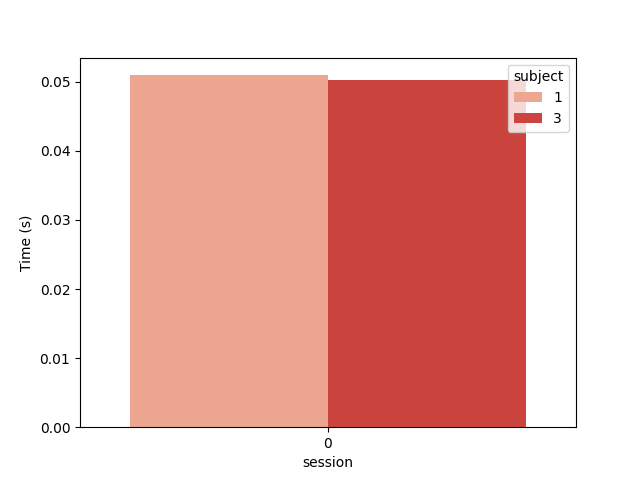
And the computation time in seconds
plt.figure()
ax = sns.barplot(data=results, y="time", x="session", hue="subject", palette="Reds")
ax.set_ylabel("Time (s)")
plt.show()
Total running time of the script: ( 0 minutes 38.890 seconds)
Estimated memory usage: 11 MB