API and Main Concepts#
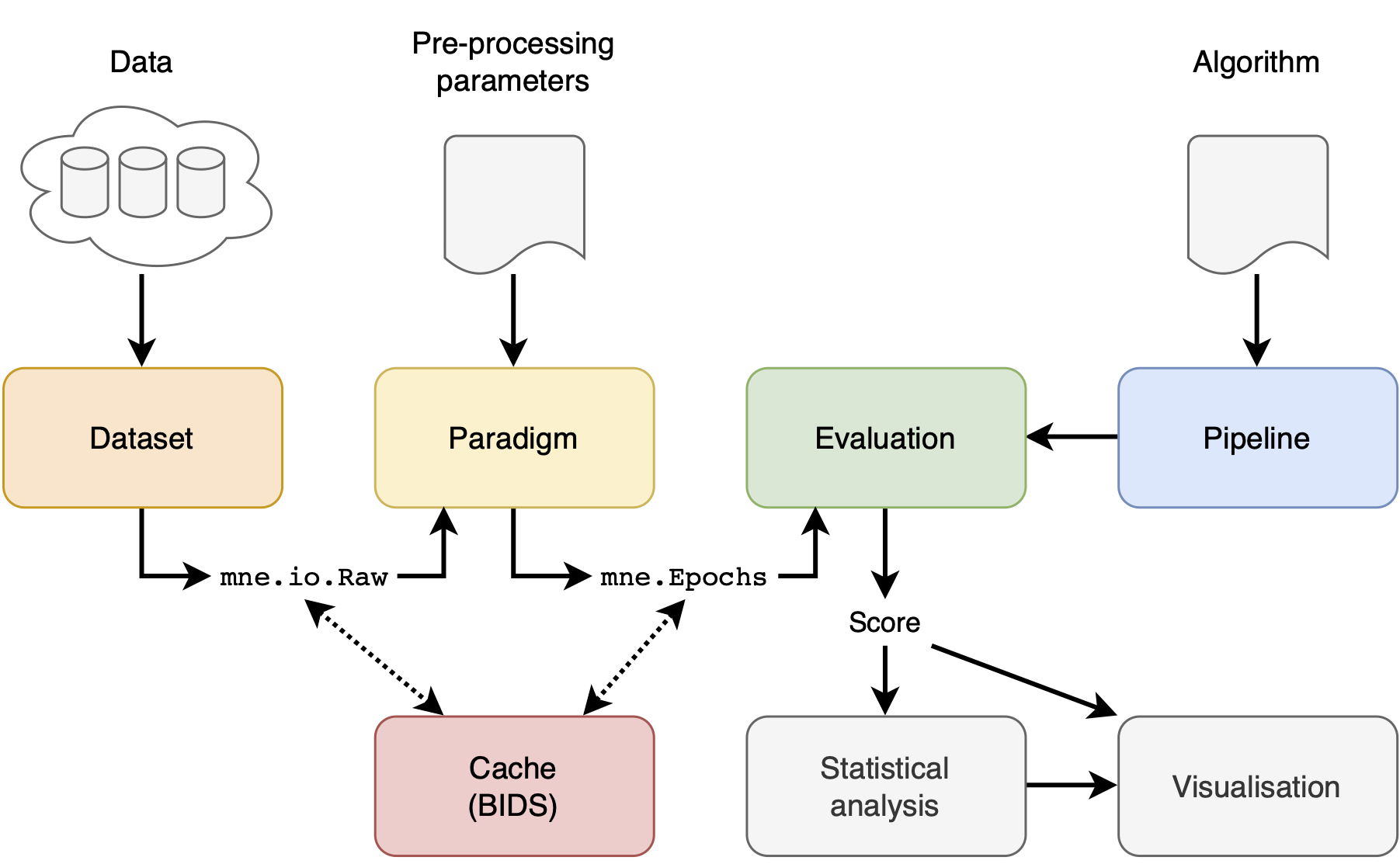
There are 4 main concepts in the MOABB: the datasets, the paradigm, the evaluation, and the pipelines. In addition, we offer statistical, visualization, utilities to simplify the workflow.
And if you want to just run the benchmark, you can use our benchmark module that wraps all the steps in a single function.
Datasets#
A dataset handles and abstracts low-level access to the data. The dataset will read data stored locally, in the format in which they have been downloaded, and will convert them into an MNE raw object. There are options to pool all the different recording sessions per subject or to evaluate them separately.
Motor Imagery Datasets#
|
Alex Motor Imagery dataset. |
BNCI 2014-001 Motor Imagery dataset. |
|
BNCI 2014-002 Motor Imagery dataset. |
|
BNCI 2014-004 Motor Imagery dataset. |
|
BNCI 2015-001 Motor Imagery dataset. |
|
BNCI 2015-004 Motor Imagery dataset. |
|
|
Motor Imagery dataset from Cho et al 2017. |
Class for Dreyer2023 dataset management. |
|
Class for Dreyer2023A dataset management. |
|
Class for Dreyer2023B dataset management. |
|
Class for Dreyer2023C dataset management. |
|
|
BMI/OpenBMI dataset for MI. |
Munich Motor Imagery dataset. |
|
|
Motor Imagery ataset from Ofner et al 2017. |
|
Physionet Motor Imagery dataset. |
High-gamma dataset described in Schirrmeister et al. 2017. |
|
|
Motor Imagey Dataset from Shin et al 2017. |
|
Mental Arithmetic Dataset from Shin et al 2017. |
Motor Imagery dataset from Weibo et al 2014. |
|
|
Motor Imagery dataset from Zhou et al 2016. |
|
Motor Imagery dataset from Stieger et al. 2021 [R9d4dafd3db21-1]. |
|
Dataset [R5c8c28f00274-1] from the study on motor imagery [R5c8c28f00274-2]. |
|
Motor Imagery dataset from BEETL Competition - Dataset A. |
|
Motor Imagery dataset from BEETL Competition - Dataset B. |
ERP/P300 Datasets#
|
P300 dataset BI2012 from a "Brain Invaders" experiment. |
|
P300 dataset BI2013a from a "Brain Invaders" experiment. |
|
P300 dataset BI2014a from a "Brain Invaders" experiment. |
|
P300 dataset BI2014b from a "Brain Invaders" experiment. |
|
P300 dataset BI2015a from a "Brain Invaders" experiment. |
|
P300 dataset BI2015b from a "Brain Invaders" experiment. |
|
Dataset of an EEG-based BCI experiment in Virtual Reality using P300. |
BNCI 2014-008 P300 dataset. |
|
BNCI 2014-009 P300 dataset. |
|
BNCI 2015-003 P300 dataset. |
|
Visual P300 dataset recorded in Virtual Reality (VR) game Raccoons versus Demons. |
|
|
P300 dataset from Hoffmann et al 2008. |
|
Learning from label proportions for a visual matrix speller (ERP) dataset from Hübner et al 2017 [R0a211c89d39d-1]. |
|
Mixture of LLP and EM for a visual matrix speller (ERP) dataset from Hübner et al 2018 [R8f30fc0d0ace-1]. |
|
BMI/OpenBMI dataset for P300. |
|
P300 dataset from initial spot study. |
ERN events of the ERP CORE dataset by Kappenman et al. 2020. |
|
LRP events of the ERP CORE dataset by Kappenman et al. 2020. |
|
MMN events of the ERP CORE dataset by Kappenman et al. 2020. |
|
N2pc events of the ERP CORE dataset by Kappenman et al. 2020. |
|
N170 events of the ERP CORE dataset by Kappenman et al. 2020. |
|
N400 events of the ERP CORE dataset by Kappenman et al. 2020. |
|
P3 events of the ERP CORE dataset by Kappenman et al. 2020. |
|
|
MOABB class for BrainForm event-related potentials (ERP) dataset. |
Class for Kojima2024A dataset management. |
|
|
Class for Kojima2024B dataset management. |
SSVEP Datasets#
SSVEP Exo dataset. |
|
SSVEP Nakanishi 2015 dataset. |
|
|
SSVEP Wang 2016 dataset. |
|
SSVEP MAMEM 1 dataset. |
|
SSVEP MAMEM 2 dataset. |
|
SSVEP MAMEM 3 dataset. |
|
BMI/OpenBMI dataset for SSVEP. |
c-VEP Datasets#
c-VEP dataset from Thielen et al. (2015). |
|
c-VEP dataset from Thielen et al. (2021). |
|
|
c-VEP and Burst-VEP dataset from Castillos et al. (2023). |
|
c-VEP and Burst-VEP dataset from Castillos et al. (2023). |
|
c-VEP and Burst-VEP dataset from Castillos et al. (2023). |
|
c-VEP and Burst-VEP dataset from Castillos et al. (2023). |
Resting State Datasets#
Passive Head Mounted Display with Music Listening dataset [Rf4e3a2801e75-1]. |
|
Neuroergonomic 2021 dataset. |
|
Alphawaves dataset |
Compound Datasets#
A selection of subject from BI2014a with AUC < 0.7 with pipeline: ERPCovariances(estimator="lwf"), MDM(metric="riemann") |
|
A selection of subject from BI2014b with AUC < 0.7 with pipeline: ERPCovariances(estimator="lwf"), MDM(metric="riemann") |
|
A selection of subject from BI2015a with AUC < 0.7 with pipeline: ERPCovariances(estimator="lwf"), MDM(metric="riemann") |
|
A selection of subject from BI2015b with AUC < 0.7 with pipeline: ERPCovariances(estimator="lwf"), MDM(metric="riemann") |
|
A selection of subject from Cattan2019_VR with AUC < 0.7 with pipeline: ERPCovariances(estimator="lwf"), MDM(metric="riemann") |
|
|
Subjects from braininvaders datasets with AUC < 0.7 with pipeline: ERPCovariances(estimator="lwf"), MDM(metric="riemann") |
Base & Utils#
|
Abstract Moabb BaseDataset. |
|
Abstract BIDS dataset class. |
|
Generic local/private BIDS datasets. |
|
Configuration for caching of datasets. |
|
Fake Dataset for test purpose. |
|
Fake Cattan2019_VR dataset for test purpose. |
|
Get path to local copy of given dataset URL. |
|
Download file from url to specified path. |
|
Wrapper for HTTP request. |
|
List all the files associated with a given article. |
|
Returns a dict associating figshare file id to MD5 hash. |
|
Returns a dict associating filename to figshare file id. |
|
Returns a dict associating figshare file id to filename. |
|
Returns a list of datasets that match a given criteria. |
|
Given a list of dataset instances return a list of channels shared by all datasets. |
|
Plots all the MOABB datasets in one figure, distributed on a grid. |
|
Plots all the MOABB datasets in one figure, grouped in one cluster. |
Paradigm#
A paradigm defines how the raw data will be converted to trials ready to be processed by a decoding algorithm. This is a function of the paradigm used, i.e. in motor imagery one can have two-class, multi-class, or continuous paradigms; similarly, different preprocessing is necessary for ERP vs ERD paradigms.
Motor Imagery Paradigms#
|
N-class motor imagery. |
|
Motor Imagery for left hand/right hand classification. |
|
Filter Bank Motor Imagery for left hand/right hand classification. |
|
Filter bank n-class motor imagery. |
P300 Paradigms#
|
Single Bandpass filter P300. |
|
P300 for Target/NonTarget classification. |
SSVEP Paradigms#
|
Single bandpass filter SSVEP. |
|
Filtered bank n-class SSVEP paradigm. |
c-VEP Paradigms#
|
Single bandpass c-VEP paradigm for epoch-level decoding. |
|
Filterbank c-VEP paradigm for epoch-level decoding. |
Resting state Paradigms#
|
Adapter to the P300 paradigm for resting state experiments. |
Fixed Interval Windows Processings#
|
Fixed interval windows processing. |
Filter bank fixed interval windows processing. |
Base & Utils#
|
Base Motor imagery paradigm. |
|
Single Bandpass filter motor Imagery. |
|
Filter Bank MI. |
|
Base P300 paradigm. |
|
Base SSVEP Paradigm. |
Base class for fixed interval windows processing. |
|
|
Base class for paradigms. |
|
Base Processing. |
Evaluations#
An evaluation defines how we go from trials per subject and session to a generalization statistic (AUC score, f-score, accuracy, etc) – it can be either within-recording-session accuracy, across-session within-subject accuracy, across-subject accuracy, or other transfer learning settings.
|
Performance evaluation within session (k-fold cross-validation) |
|
Cross-session performance evaluation. |
|
Cross-subject evaluation performance. |
|
Data splitter for within session evaluation. |
|
Data splitter for cross session evaluation. |
|
Data splitter for cross subject evaluation. |
Base & Utils#
|
Base class that defines necessary operations for an evaluation. |
Pipelines#
Pipeline defines all steps required by an algorithm to obtain predictions. Pipelines are typically a chain of sklearn compatible transformers and end with a sklearn compatible estimator. See Pipelines for more info.
LogVariance transformer. |
|
|
Transformer to scale sampling frequency. |
Prepare FilterBank SSVEP EEG signal for estimating extended covariances. |
|
|
Dataset augmentation methods in a higher dimensional space. |
Function to standardize the X raw data. |
|
|
Weighted Tikhonov-regularized CSP as described in Lotte and Guan 2011. |
|
Classifier based on Canonical Correlation Analysis for SSVEP. |
|
Classifier based on the Task-Related Component Analysis method [Rdaa19db1b521-1] for SSVEP. |
|
Classifier based on MsetCCA for SSVEP. |
Statistics, visualization and utilities#
Once an evaluation has been run, the raw results are returned as a DataFrame. This can be further processed via the following commands to generate some basic visualization and statistical comparisons:
Plotting#
|
Plot scores for all pipelines and all datasets |
|
Generate a figure with a paired plot. |
|
Significance matrix to compare pipelines. |
|
Meta-analysis to compare two algorithms across several datasets. |
|
Plot a bubble plot for a dataset. |
Statistics#
Compute differences between pipelines across datasets. |
|
Compute meta-analysis statistics from results dataframe. |
|
|
Combine effects for meta-analysis statistics. |
|
Combine p-values for meta-analysis statistics. |
Prepare results dataframe for computing statistics. |
Utils#
|
Set log level. |
|
Set the seed for random, numpy. |
|
Set the download directory if required to change from default mne path. |
|
Shortcut for the method |
Benchmark#
The benchmark module wraps all the steps in a single function. It downloads the data, runs the benchmark, and returns the results. It is the easiest way to run a benchmark.
from moabb import benchmark
results = benchmark(
pipelines="./pipelines",
evaluations=["WithinSession"],
paradigms=["LeftRightImagery"],
include_datasets=[BNCI2014_001(), PhysionetMI()],
exclude_datasets=None,
results="./results/",
overwrite=True,
plot=True,
output="./benchmark/",
n_jobs=-1,
)
|
Run benchmarks for selected pipelines and datasets. |