Note
Click here to download the full example code
Fixed interval windows processing#
This example shows how to process a dataset using the
moabb.paradigms.FixedIntervalWindowsProcessing paradigm. This paradigm
creates epochs at fixed intervals, ignoring the stim
channel and events of the datasets. Therefore, it is
compatible with all the datasets. Unfortunately,
this paradigm is not compatible with the MOABB evaluation
framework. However, it can be used to process datasets
for unsupervised algorithms.
In this example, we will use the Zhou2016 dataset because it is relatively small and can be downloaded quickly.
# Authors: Pierre Guetschel <pierre.guetschel@gmail.com>
#
# License: BSD (3-clause)
import matplotlib.pyplot as plt
import mne
import numpy as np
from moabb import set_log_level
from moabb.datasets import Zhou2016
from moabb.paradigms import FixedIntervalWindowsProcessing, MotorImagery
set_log_level("info")
Process a dataset#
To process a dataset with
moabb.paradigms.FixedIntervalWindowsProcessing , you can use the
method as with every other paradigm. The only additional parameters are
length, stride, start_offset, and stop_offset. They are
all parametrised in seconds. length is the length of the epochs,
stride is the time between the onset of two consecutive epochs,
start_offset is the offset between each run start and their first
epoch, and stop_offset is the offset between each run start and their
last epoch. The default values are length=5, stride=10,
start_offset=0, and stop_offset=None (i.e. end of the run).
An example usage of moabb.paradigms.FixedIntervalWindowsProcessing
with the moabb.datasets.Zhou2016 dataset:
dataset = Zhou2016()
processing = FixedIntervalWindowsProcessing(
# new parameters:
length=100,
stride=50,
start_offset=300,
stop_offset=900, # we epoch 10 minutes per run, starting at 5 minutes (i.e. 300 seconds)
# parameters common with other paradigms:
resample=100,
fmin=7,
fmax=45,
baseline=None,
channels=None,
)
X, labels, metadata = processing.get_data(dataset=dataset, subjects=[1])
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 10 events (all good), 0 – 100 s (baseline off), ~26.7 MB, data loaded,
'Window': 10>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 10 events (all good), 0 – 100 s (baseline off), ~26.7 MB, data loaded,
'Window': 10>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 10 events (all good), 0 – 100 s (baseline off), ~26.7 MB, data loaded,
'Window': 10>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 10 events (all good), 0 – 100 s (baseline off), ~26.7 MB, data loaded,
'Window': 10>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 10 events (all good), 0 – 100 s (baseline off), ~26.7 MB, data loaded,
'Window': 10>
warn(f"warnEpochs {epochs}")
/home/runner/work/moabb/moabb/moabb/datasets/preprocessing.py:278: UserWarning: warnEpochs <Epochs | 10 events (all good), 0 – 100 s (baseline off), ~26.7 MB, data loaded,
'Window': 10>
warn(f"warnEpochs {epochs}")
In this dataset, there are three sessions per subject and two runs per session:
subjects: [1]
sessions: ['0' '1' '2']
runs: ['0' '1']
We expect to obtained (stop_offset - start_offset - length) / stride;
i.e. (900-300-100)/50=10 epochs per run. Here we have 3*2=6 runs.
And indeed, we obtain
a total of 6*10=60 epochs:
print(f"Number of epochs: {len(X)}")
Number of epochs: 60
Note
To apply a bank of bandpass filters, you can use the
moabb.paradigms.FilterBankFixedIntervalWindowsProcessing
paradigm instead.
Print the events#
We can print the position of the created epochs within the run next to
the original events of the dataset. For this, we will first instantiate
a moabb.paradigms.MotorImagery paradigm to recover the original
events of the dataset:
paradigm = MotorImagery(
resample=100,
fmin=7,
fmax=45,
baseline=None,
channels=None,
)
Then, we can recover the events of both paradigms using the
_get_events_pipeline method:
events_pipeline_dataset = paradigm._get_events_pipeline(dataset)
events_pipeline_fixed = processing._get_events_pipeline(dataset)
raw = dataset.get_data(subjects=[1])[1]["0"]["0"]
events_dataset = events_pipeline_dataset.transform(raw)
events_fixed = events_pipeline_fixed.transform(raw)
events = np.concatenate([events_dataset, events_fixed])
event_id = dict(**paradigm.used_events(dataset), **processing.used_events(dataset))
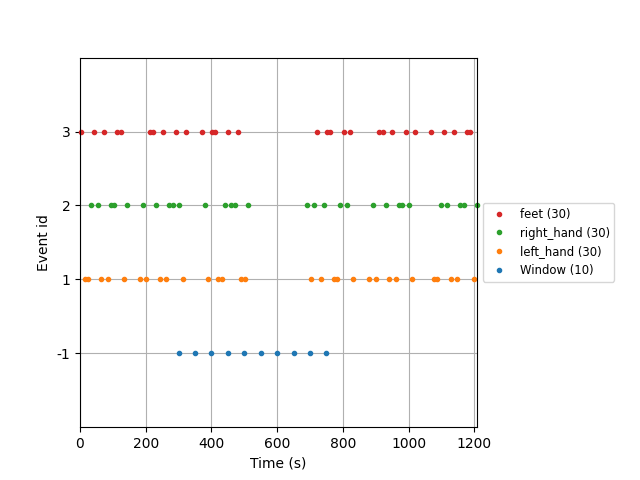
Finally, we can plot the events. The artificial events created by
moabb.paradigms.FixedIntervalWindowsProcessing are named
"Windows":
fig = mne.viz.plot_events(
events,
sfreq=raw.info["sfreq"],
event_id=event_id,
)
fig.subplots_adjust(right=0.7)
plt.show()
/home/runner/work/moabb/moabb/examples/plot_fixed_interval_windows.py:122: UserWarning: This figure was using a layout engine that is incompatible with subplots_adjust and/or tight_layout; not calling subplots_adjust.
fig.subplots_adjust(right=0.7)
We can see that the epochs were effectively created at a fixed interval every 50 seconds between 300 and 900 seconds, and ignoring the original events of the dataset.
Total running time of the script: ( 0 minutes 11.490 seconds)
Estimated memory usage: 387 MB