Note
Go to the end to download the full example code.
Within Session P300#
This example shows how to perform a within session analysis on three different P300 datasets.
We will compare two pipelines :
Riemannian geometry
XDAWN with Linear Discriminant Analysis
We will use the P300 paradigm, which uses the AUC as metric.
# Authors: Pedro Rodrigues <pedro.rodrigues01@gmail.com>
#
# License: BSD (3-clause)
import warnings
import matplotlib.pyplot as plt
import seaborn as sns
from mne.decoding import Vectorizer
from pyriemann.estimation import Xdawn, XdawnCovariances
from pyriemann.tangentspace import TangentSpace
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.pipeline import make_pipeline
import moabb
from moabb.datasets import BNCI2014_009
from moabb.evaluations import WithinSessionEvaluation
from moabb.paradigms import P300
getting rid of the warnings about the future
warnings.simplefilter(action="ignore", category=FutureWarning)
warnings.simplefilter(action="ignore", category=RuntimeWarning)
moabb.set_log_level("info")
Create Pipelines#
Pipelines must be a dict of sklearn pipeline transformer.
pipelines = {}
We have to do this because the classes are called ‘Target’ and ‘NonTarget’ but the evaluation function uses a LabelEncoder, transforming them to 0 and 1
labels_dict = {"Target": 1, "NonTarget": 0}
pipelines["RG+LDA"] = make_pipeline(
XdawnCovariances(
nfilter=2, classes=[labels_dict["Target"]], estimator="lwf", xdawn_estimator="scm"
),
TangentSpace(),
LDA(solver="lsqr", shrinkage="auto"),
)
pipelines["Xdw+LDA"] = make_pipeline(
Xdawn(nfilter=2, estimator="scm"), Vectorizer(), LDA(solver="lsqr", shrinkage="auto")
)
Evaluation#
We define the paradigm (P300) and use all three datasets available for it. The evaluation will return a DataFrame containing a single AUC score for each subject / session of the dataset, and for each pipeline.
Results are saved into the database, so that if you add a new pipeline, it will not run again the evaluation unless a parameter has changed. Results can be overwritten if necessary.
paradigm = P300(resample=128)
dataset = BNCI2014_009()
dataset.subject_list = dataset.subject_list[:2]
datasets = [dataset]
overwrite = True # set to True if we want to overwrite cached results
evaluation = WithinSessionEvaluation(
paradigm=paradigm, datasets=datasets, suffix="examples", overwrite=overwrite
)
results = evaluation.process(pipelines)
BNCI2014-009-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
BNCI2014-009-WithinSession: 50%|█████ | 1/2 [00:08<00:08, 8.20s/it]
BNCI2014-009-WithinSession: 100%|██████████| 2/2 [00:16<00:00, 8.46s/it]
BNCI2014-009-WithinSession: 100%|██████████| 2/2 [00:16<00:00, 8.42s/it]
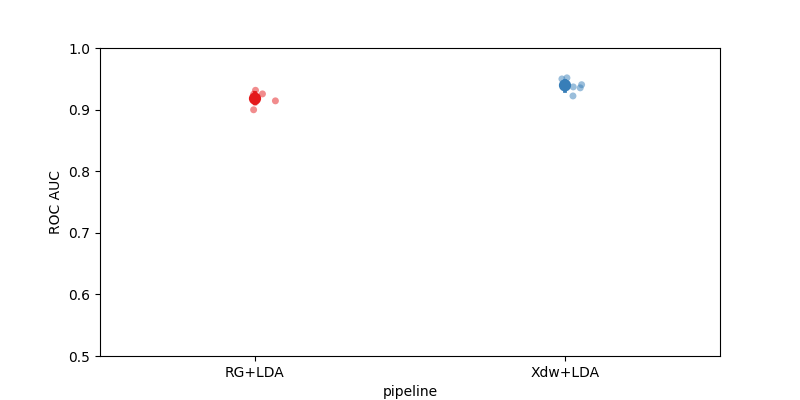
Plot Results#
Here we plot the results to compare the two pipelines
fig, ax = plt.subplots(facecolor="white", figsize=[8, 4])
sns.stripplot(
data=results,
y="score",
x="pipeline",
ax=ax,
jitter=True,
alpha=0.5,
zorder=1,
palette="Set1",
)
sns.pointplot(data=results, y="score", x="pipeline", ax=ax, palette="Set1")
ax.set_ylabel("ROC AUC")
ax.set_ylim(0.5, 1)
plt.show()
Total running time of the script: (0 minutes 19.755 seconds)
Estimated memory usage: 310 MB