Note
Go to the end to download the full example code.
Within Session Motor Imagery with Learning Curve#
This example shows how to perform a within session motor imagery analysis on the very popular dataset 2a from the BCI competition IV.
We will compare two pipelines :
CSP + LDA
Riemannian Geometry + Logistic Regression
We will use the LeftRightImagery paradigm. This will restrict the analysis to two classes (left- vs right-hand) and use AUC as metric.
# Original author: Alexandre Barachant <alexandre.barachant@gmail.com>
# Learning curve modification: Jan Sosulski
#
# License: BSD (3-clause)
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from mne.decoding import CSP
from pyriemann.estimation import Covariances
from pyriemann.tangentspace import TangentSpace
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import make_pipeline
import moabb
from moabb.datasets import BNCI2014_001
from moabb.evaluations import WithinSessionEvaluation
from moabb.paradigms import LeftRightImagery
moabb.set_log_level("info")
Create Pipelines#
Pipelines must be a dict of sklearn pipeline transformer.
The CSP implementation from MNE is used. We selected 8 CSP components, as usually done in the literature.
The Riemannian geometry pipeline consists in covariance estimation, tangent space mapping and finally a logistic regression for the classification.
pipelines = {}
pipelines["CSP+LDA"] = make_pipeline(
CSP(n_components=8), LDA(solver="lsqr", shrinkage="auto")
)
pipelines["RG+LR"] = make_pipeline(
Covariances(), TangentSpace(), LogisticRegression(solver="lbfgs")
)
Evaluation#
We define the paradigm (LeftRightImagery) and the dataset (BNCI2014_001). The evaluation will return a DataFrame containing a single AUC score for each subject / session of the dataset, and for each pipeline.
Results are saved into the database, so that if you add a new pipeline, it will not run again the evaluation unless a parameter has changed. Results can be overwritten if necessary.
paradigm = LeftRightImagery()
dataset = BNCI2014_001()
dataset.subject_list = dataset.subject_list[:1]
datasets = [dataset]
overwrite = True # set to True if we want to overwrite cached results
# Evaluate for a specific number of training samples per class
data_size = dict(policy="per_class", value=np.array([5, 10, 30, 50]))
# When the training data is sparse, perform more permutations than when we have a lot of data
n_perms = np.floor(np.geomspace(20, 2, len(data_size["value"]))).astype(int)
evaluation = WithinSessionEvaluation(
paradigm=paradigm,
datasets=datasets,
suffix="examples",
overwrite=overwrite,
data_size=data_size,
n_perms=n_perms,
)
results = evaluation.process(pipelines)
print(results.head())
BNCI2014-001-WithinSession: 0%| | 0/1 [00:00<?, ?it/s]
BNCI2014-001-WithinSession: 100%|██████████| 1/1 [00:14<00:00, 14.49s/it]
BNCI2014-001-WithinSession: 100%|██████████| 1/1 [00:14<00:00, 14.49s/it]
score time samples ... n_sessions dataset pipeline
0 0.704762 0.036649 10.0 ... 2 BNCI2014-001 CSP+LDA
1 0.966667 0.098719 20.0 ... 2 BNCI2014-001 CSP+LDA
2 0.938095 0.131995 60.0 ... 2 BNCI2014-001 CSP+LDA
3 0.980952 0.243238 100.0 ... 2 BNCI2014-001 CSP+LDA
4 0.814286 0.054529 10.0 ... 2 BNCI2014-001 CSP+LDA
[5 rows x 11 columns]
Plot Results#
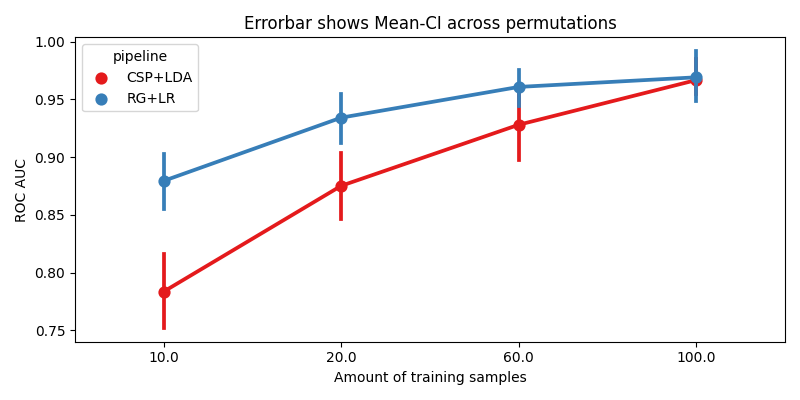
We plot the accuracy as a function of the number of training samples, for each pipeline
fig, ax = plt.subplots(facecolor="white", figsize=[8, 4])
n_subs = len(dataset.subject_list)
if n_subs > 1:
r = results.groupby(["pipeline", "subject", "data_size"]).mean().reset_index()
else:
r = results
sns.pointplot(data=r, x="data_size", y="score", hue="pipeline", ax=ax, palette="Set1")
errbar_meaning = "subjects" if n_subs > 1 else "permutations"
title_str = f"Errorbar shows Mean-CI across {errbar_meaning}"
ax.set_xlabel("Amount of training samples")
ax.set_ylabel("ROC AUC")
ax.set_title(title_str)
fig.tight_layout()
plt.show()
Total running time of the script: (0 minutes 32.755 seconds)
Estimated memory usage: 572 MB