Note
Go to the end to download the full example code.
Select Electrodes and Resampling#
Within paradigm, it is possible to restrict analysis only to a subset of electrodes and to resample to a specific sampling rate. There is also a utility function to select common electrodes shared between datasets. This tutorial demonstrates how to use this functionality.
# Authors: Sylvain Chevallier <sylvain.chevallier@uvsq.fr>
#
# License: BSD (3-clause)
import matplotlib.pyplot as plt
from mne.decoding import CSP
from pyriemann.estimation import Covariances
from pyriemann.tangentspace import TangentSpace
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.linear_model import LogisticRegression as LR
from sklearn.pipeline import make_pipeline
import moabb.analysis.plotting as moabb_plt
from moabb.datasets import BNCI2014_001, Zhou2016
from moabb.datasets.utils import find_intersecting_channels
from moabb.evaluations import WithinSessionEvaluation
from moabb.paradigms import LeftRightImagery
Datasets#
Load 2 subjects of BNCI 2014-004 and Zhou2016 datasets, with 2 sessions each
subj = [1, 2]
datasets = [Zhou2016(), BNCI2014_001()]
for d in datasets:
d.subject_list = subj
Paradigm#
Restrict further analysis to specified channels, here C3, C4, and Cz. Also, use a specific resampling. In this example, all datasets are set to 200 Hz.
paradigm = LeftRightImagery(channels=["C3", "C4", "Cz"], resample=200.0)
Evaluation#
The evaluation is conducted on with CSP+LDA, only on the 3 electrodes, with a sampling rate of 200 Hz.
evaluation = WithinSessionEvaluation(paradigm=paradigm, datasets=datasets)
csp_lda = make_pipeline(CSP(n_components=2), LDA())
ts_lr = make_pipeline(
Covariances(estimator="oas"), TangentSpace(metric="riemann"), LR(C=1.0)
)
results = evaluation.process({"csp+lda": csp_lda, "ts+lr": ts_lr})
print(results.head())
Zhou2016-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
Zhou2016-WithinSession: 50%|█████ | 1/2 [00:02<00:02, 2.63s/it]
Zhou2016-WithinSession: 100%|██████████| 2/2 [00:04<00:00, 2.38s/it]
Zhou2016-WithinSession: 100%|██████████| 2/2 [00:04<00:00, 2.42s/it]
BNCI2014-001-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
BNCI2014-001-WithinSession: 50%|█████ | 1/2 [00:03<00:03, 3.48s/it]
BNCI2014-001-WithinSession: 100%|██████████| 2/2 [00:06<00:00, 3.49s/it]
BNCI2014-001-WithinSession: 100%|██████████| 2/2 [00:06<00:00, 3.49s/it]
score time samples subject ... channels n_sessions dataset pipeline
0 0.853914 0.067811 119.0 1 ... 3 3 Zhou2016 ts+lr
1 0.916000 0.056730 100.0 1 ... 3 3 Zhou2016 ts+lr
2 0.946000 0.056742 100.0 1 ... 3 3 Zhou2016 ts+lr
3 0.926000 0.054432 100.0 2 ... 3 3 Zhou2016 ts+lr
4 0.822222 0.049617 90.0 2 ... 3 3 Zhou2016 ts+lr
[5 rows x 9 columns]
Electrode Selection#
It is possible to select the electrodes that are shared by all datasets using the find_intersecting_channels function. Datasets that have 0 overlap with others are discarded. It returns the set of common channels, as well as the list of datasets with valid channels.
electrodes, datasets = find_intersecting_channels(datasets)
evaluation = WithinSessionEvaluation(
paradigm=paradigm, datasets=datasets, overwrite=True, suffix="resample"
)
results = evaluation.process({"csp+lda": csp_lda, "ts+lr": ts_lr})
print(results.head())
Zhou2016-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
Zhou2016-WithinSession: 50%|█████ | 1/2 [00:02<00:02, 2.64s/it]
Zhou2016-WithinSession: 100%|██████████| 2/2 [00:04<00:00, 2.44s/it]
Zhou2016-WithinSession: 100%|██████████| 2/2 [00:04<00:00, 2.47s/it]
BNCI2014-001-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
BNCI2014-001-WithinSession: 50%|█████ | 1/2 [00:03<00:03, 3.48s/it]
BNCI2014-001-WithinSession: 100%|██████████| 2/2 [00:06<00:00, 3.44s/it]
BNCI2014-001-WithinSession: 100%|██████████| 2/2 [00:06<00:00, 3.44s/it]
score time samples subject ... channels n_sessions dataset pipeline
0 0.848611 0.067350 119.0 1 ... 3 3 Zhou2016 ts+lr
1 0.922000 0.059805 100.0 1 ... 3 3 Zhou2016 ts+lr
2 0.938000 0.055341 100.0 1 ... 3 3 Zhou2016 ts+lr
3 0.922000 0.055319 100.0 2 ... 3 3 Zhou2016 ts+lr
4 0.849383 0.047388 90.0 2 ... 3 3 Zhou2016 ts+lr
[5 rows x 9 columns]
Plot Results#
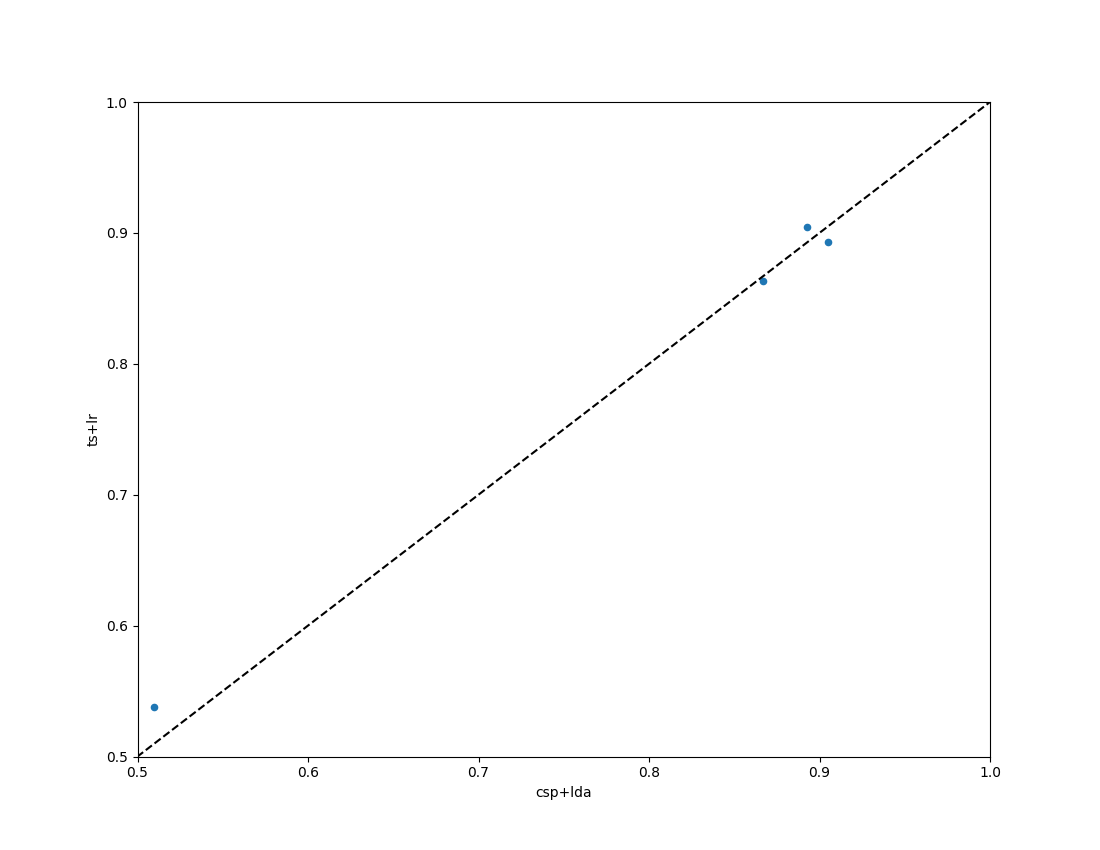
Compare the obtained results with the two pipelines, CSP+LDA and logistic regression computed in the tangent space of the covariance matrices.
fig = moabb_plt.paired_plot(results, "csp+lda", "ts+lr")
plt.show()
Total running time of the script: (0 minutes 32.742 seconds)
Estimated memory usage: 728 MB