Note
Go to the end to download the full example code.
Select Electrodes and Resampling#
Within paradigm, it is possible to restrict analysis only to a subset of electrodes and to resample to a specific sampling rate. There is also a utility function to select common electrodes shared between datasets. This tutorial demonstrates how to use this functionality.
# Authors: Sylvain Chevallier <sylvain.chevallier@uvsq.fr>
#
# License: BSD (3-clause)
import matplotlib.pyplot as plt
from mne.decoding import CSP
from pyriemann.estimation import Covariances
from pyriemann.tangentspace import TangentSpace
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.linear_model import LogisticRegression as LR
from sklearn.pipeline import make_pipeline
import moabb.analysis.plotting as moabb_plt
from moabb.datasets import BNCI2014_001, Zhou2016
from moabb.datasets.utils import find_intersecting_channels
from moabb.evaluations import WithinSessionEvaluation
from moabb.paradigms import LeftRightImagery
Datasets#
Load 2 subjects of BNCI 2014-004 and Zhou2016 datasets, with 2 sessions each
subj = [1, 2]
datasets = [Zhou2016(), BNCI2014_001()]
for d in datasets:
d.subject_list = subj
Paradigm#
Restrict further analysis to specified channels, here C3, C4, and Cz. Also, use a specific resampling. In this example, all datasets are set to 200 Hz.
paradigm = LeftRightImagery(channels=["C3", "C4", "Cz"], resample=200.0)
Evaluation#
The evaluation is conducted on with CSP+LDA, only on the 3 electrodes, with a sampling rate of 200 Hz.
evaluation = WithinSessionEvaluation(paradigm=paradigm, datasets=datasets)
csp_lda = make_pipeline(CSP(n_components=2), LDA())
ts_lr = make_pipeline(
Covariances(estimator="oas"), TangentSpace(metric="riemann"), LR(C=1.0)
)
results = evaluation.process({"csp+lda": csp_lda, "ts+lr": ts_lr})
print(results.head())
Zhou2016-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
Zhou2016-WithinSession: 50%|█████ | 1/2 [00:03<00:03, 3.59s/it]
Zhou2016-WithinSession: 100%|██████████| 2/2 [00:06<00:00, 3.41s/it]
Zhou2016-WithinSession: 100%|██████████| 2/2 [00:06<00:00, 3.43s/it]
BNCI2014-001-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
BNCI2014-001-WithinSession: 50%|█████ | 1/2 [00:05<00:05, 5.18s/it]
BNCI2014-001-WithinSession: 100%|██████████| 2/2 [00:09<00:00, 4.81s/it]
BNCI2014-001-WithinSession: 100%|██████████| 2/2 [00:09<00:00, 4.86s/it]
score time ... pipeline codecarbon_task_name
0 0.847601 0.047893 ... ts+lr 2dd18d9e-58ce-4839-8778-bda96f315583
1 0.922000 0.043081 ... ts+lr 651199d9-3ec5-41fc-bd41-b7743ba1d9ed
2 0.952000 0.045720 ... ts+lr cf73a530-c110-43c5-99b4-a10c62713349
3 0.916000 0.045387 ... ts+lr 49a2d4e5-4588-41d9-a720-d115b6f3bdaf
4 0.846914 0.041111 ... ts+lr f9d1e5f7-2929-45e5-9c2e-a6ee71f866c9
[5 rows x 11 columns]
Electrode Selection#
It is possible to select the electrodes that are shared by all datasets using the find_intersecting_channels function. Datasets that have 0 overlap with others are discarded. It returns the set of common channels, as well as the list of datasets with valid channels.
electrodes, datasets = find_intersecting_channels(datasets)
evaluation = WithinSessionEvaluation(
paradigm=paradigm, datasets=datasets, overwrite=True, suffix="resample"
)
results = evaluation.process({"csp+lda": csp_lda, "ts+lr": ts_lr})
print(results.head())
Zhou2016-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
Zhou2016-WithinSession: 50%|█████ | 1/2 [00:03<00:03, 3.41s/it]
Zhou2016-WithinSession: 100%|██████████| 2/2 [00:07<00:00, 3.61s/it]
Zhou2016-WithinSession: 100%|██████████| 2/2 [00:07<00:00, 3.58s/it]
BNCI2014-001-WithinSession: 0%| | 0/2 [00:00<?, ?it/s]
BNCI2014-001-WithinSession: 50%|█████ | 1/2 [00:05<00:05, 5.30s/it]
BNCI2014-001-WithinSession: 100%|██████████| 2/2 [00:10<00:00, 4.96s/it]
BNCI2014-001-WithinSession: 100%|██████████| 2/2 [00:10<00:00, 5.01s/it]
score time ... pipeline codecarbon_task_name
0 0.839141 0.051255 ... ts+lr fc7ba9ac-3007-4812-b0aa-e5321c7f5402
1 0.912000 0.041649 ... ts+lr 6eb79699-8ecc-4e6b-9720-a6c37daa6240
2 0.950000 0.052880 ... ts+lr a3752541-cbe6-4a23-907e-2a714f32517a
3 0.930000 0.056838 ... ts+lr 2b7f8dbc-1357-4f29-b8d9-10c79aa9ef1b
4 0.827160 0.051731 ... ts+lr ac2cc52f-a866-4c5c-ae11-e55494798f05
[5 rows x 11 columns]
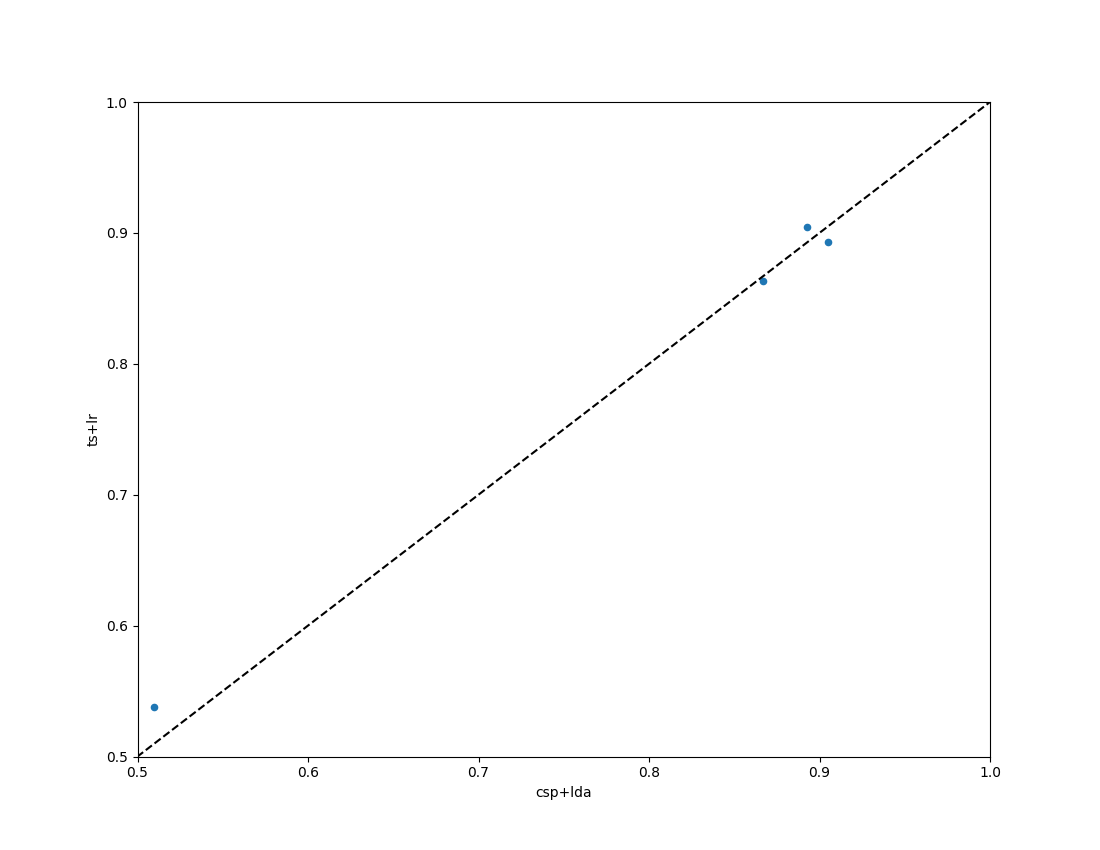
Plot Results#
Compare the obtained results with the two pipelines, CSP+LDA and logistic regression computed in the tangent space of the covariance matrices.
fig = moabb_plt.paired_plot(results, "csp+lda", "ts+lr")
plt.show()
Total running time of the script: (0 minutes 45.370 seconds)